Step-by-step guide on using Random Forests to handle missing data
Missing value imputation is an ever-old question in data science and machine learning. Techniques go from the simple mean/median imputation to more sophisticated methods based on machine learning. How much of an impact approach selection has on the final results? As it turns out, a lot.
Let’s get a couple of things straight — missing value imputation is domain-specific more often than not. For example, a dataset might contain missing values because a customer isn’t using some service, so imputation would be the wrong thing to do.
Further, simple techniques like mean/median/mode imputation often don’t work well. And it’s easy to reason why. Extremes can influence average values in the dataset, the mean in particular. Also, filling 10% or more of the data with the same value doesn’t sound too peachy, at least for the continuous variables.
The article is structured as follows:
- Problems with KNN imputation
- What is MissForest?
- MissForest in practice
- MissForest evaluation
- Conclusion
Problems with KNN imputation
ven some of the machine learning-based imputation techniques have issues. For example, KNN imputation is a great stepping stone from the simple average imputation but poses a couple of problems:
- You need to choose a value for K — not an issue for small datasets
- Is sensitive to outliers because it uses Euclidean distance below the surface
- Can’t be applied to categorical data, as some form of conversion to numerical representation is required
- Can be computationally expensive, but that depends on the size of your dataset
Don’t get me wrong, I would pick KNN imputation over a simple average any day, but there are still better methods.
What is MissForest?
MissForest is a machine learning-based imputation technique. It uses a Random Forest algorithm to do the task. It is based on an iterative approach, and at each iteration the generated predictions are better. You can read more about the theory of the algorithm below, as Andre Yemade great explanations and beautiful visuals:
This article aims more towards practical application, so we won’t dive too much into the theory. To summarize, MisForrest is excellent because:
- Doesn’t require extensive data preparation — as a Random forest algorithm can determine which features are important
- Doesn’t require any tuning — like K in K-Nearest Neighbors
- Doesn’t care about categorical data types — Random forest knows how to handle them
Next, we’ll dive deep into a practical example.
MissForest in practice
We’ll work with the Iris dataset for the practical part. The dataset doesn’t contain any missing values, but that’s the whole point. We will produce missing values randomly, so we can later evaluate the performance of the MissForest algorithm.
Before I forget, please install the required library by executing pip install missingpy from the Terminal.
Great! Next, let’s import Numpy and Pandas and read in the mentioned Iris dataset. We’ll also make a copy of the dataset so that we can evaluate with real values later on:
import numpy as np
import pandas as pd
iris = pd.read_csv('iris.csv')
# Keep an untouched copy for later
iris_orig = iris.copy()
iris.head()
Image by author
All right, let’s now make two lists of unique random numbers ranging from zero to the Iris dataset’s length. With some Pandas manipulation, we’ll replace the values of sepal_lengthand petal_width with NaNs, based on the index positions generated randomly:
# Generate unique lists of random integers
inds1 = list(set(np.random.randint(0, len(iris), 10)))
inds2 = list(set(np.random.randint(0, len(iris), 15)))
# Replace the values at given index position with NaNs
iris['sepal_length'] = [val if i not in inds1 else np.nan for i, val in enumerate(iris['sepal_length'])]
iris['petal_width'] = [val if i not in inds2 else np.nan for i, val in enumerate(iris['petal_width'])]
# Get count of missing values by column
iris.isnull().sum()

Image by author
As you can see, the petal_width contains only 14 missing values. That’s because the randomization process created two identical random numbers. It doesn’t pose any problem to us, as in the end, the number of missing values is arbitrary.
The next step is to, well, perform the imputation. We’ll have to remove the target variable from the picture too. Here’s how:
from missingpy import MissForest
# Make an instance and perform the imputation
imputer = MissForest()
X = iris.drop('species', axis=1)
X_imputed = imputer.fit_transform(X)
And that’s it — missing values are now imputed!
But how do we evaluate the damn thing? That’s the question we’ll answer next.
MissForest evaluation
To perform the evaluation, we’ll make use of our copied, untouched dataset. We’ll add two additional columns representing the imputed columns from the MissForest algorithm — both for sepal_length and petal_width.
We’ll then create a new dataset containing only these two columns — in the original and imputed states. Finally, we will calculate the absolute errors for further inspection.
Here’s the code:
# Add imputed values as columns to the untouched dataset
iris_orig['MF_sepal_length'] = X_imputed[:, 0]
iris_orig['MF_petal_width'] = X_imputed[:, -1]
comparison_df = iris_orig[['sepal_length', 'MF_sepal_length', 'petal_width', 'MF_petal_width']]
# Calculate absolute errors
comparison_df['ABS_ERROR_sepal_length'] = np.abs(compaison_df['sepal_length'] - comparison_df['MF_sepal_length'])
comparison_df['ABS_ERROR_petal_width'] = np.abs(compaison_df['petal_width'] - comparison_df['MF_petal_width'])
# Show only rows where imputation was performed
comparison_df.iloc[sorted([*inds1, *inds2])]
As you can see, the last line of code selects only those rows on which imputation was performed. Let’s take a look:
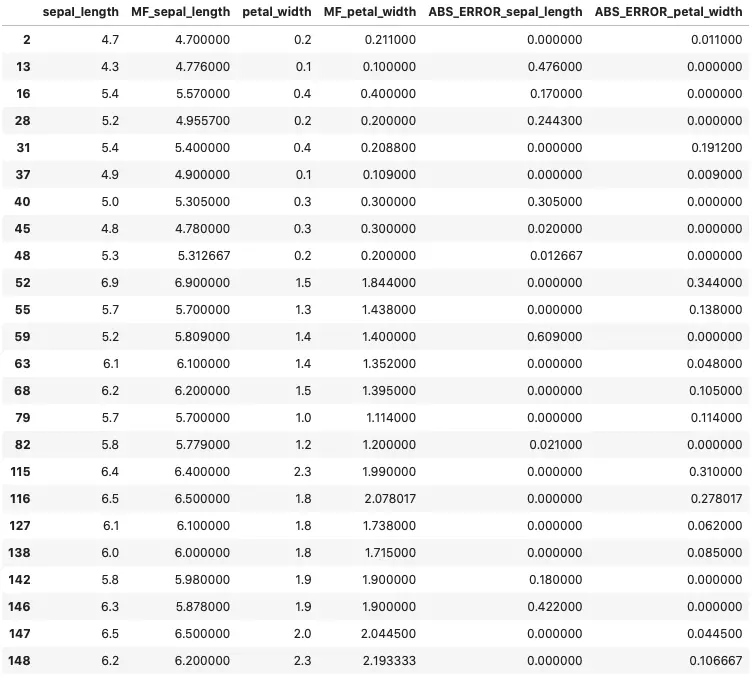
Image by author
All absolute errors are small and well within a single standard deviation from the original’s average. The imputed value looks natural if you don’t take into account the added decimal places. That can be easily fixed if necessary.
Parting words
This was a short, simple, and to the point article on missing value imputation with machine learning methods. You’ve learned why machine learning is better than the simple average in this realm and why MissForest outperforms KNN imputer.
I hope it was a good read for you. Take care.

