Machine learning foundations with R. And a bunch of other things.
I decided to start an entire series on machine learning with R. No, that doesn’t mean I’m quitting Python (God forbid), but I’ve been exploring R recently and it isn’t that bad as I initially thought. So, let start with the basics — linear regression.
This should be a fun series. I intend to cover all the major machine learning algorithms, compare the ‘weird’ parts with its _Python_alternative, and also to learn a lot of things in the process.
This article is structured as follows:
- Intro to linear regression
- Dataset introduction and loading
- Basic EDA
- Model training and evaluation
- Conclusion
Also, you can get the source code here.
We have a lot of things to cover, so let’s get started right away!
Intro to linear regression
I’ll take my chances and say that this probably isn’t your first exposure to linear regression. And it shouldn’t be, as the article won’t go in much depth with the theory. Still, we’ll start with the high-level overview of the algorithm.
Linear regression is a simple algorithm initially developed in the field of statistics. It was studied as a model for understanding relationships between input and output variables. As the name suggests, it’s a linear model, so it assumes a linear relationship between input variables and a single (continuous) output variable.
This output variable is calculated as a linear combination of the input variables.
Two main types of linear regression exist:
- Simple linear regression— when we have only one input variable
- Multiple linear regression— when there are multiple input variables
Training a linear regression model essentially adds a coefficient to each input variable — which determines how important it is. The value of input variables are then multiplied with the corresponding coefficient, and the bias (intercept) term is added to the sum. That’s essentially our predicted value.
Awesome! Linear regression has some assumption, and we as a data scientists must be aware of them:
- Linear assumption— model assumes that the relationship between variables is linear
- No noise— model assumes that the input and output variables are not noisy — so remove outliers if possible
- No collinearity— model will overfit when you have highly correlated input variables
- Normal distribution— the model will make more reliable predictions if your input and output variables are normally distributed. If that’s not the case, try using some transforms on your variables to make them more normal-looking
- Rescaled inputs— use scalers or normalizer to make more reliable predictions
And that’s it for a high-level overview. Let’s continue with the good stuff now.
Dataset introduction and loading
We’ll use the Fish Market dataset to build our model. Yes, it’s the first time I’m using it too, so you’re not alone. Download the dataset and store it in a friendly location.
Now we can start with the imports:
library(dplyr)
library(ggplot2)
library(caTools)
library(corrgram)
Put simply —dplyr is used for data manipulation, ggplot2 for visualization, caTools for train/test split, and corrgram for making neat correlation matrix plots.
We can now read in the dataset and check how do the first couple of rows look like:
df <- read.csv('data/Fish.csv')
head(df)
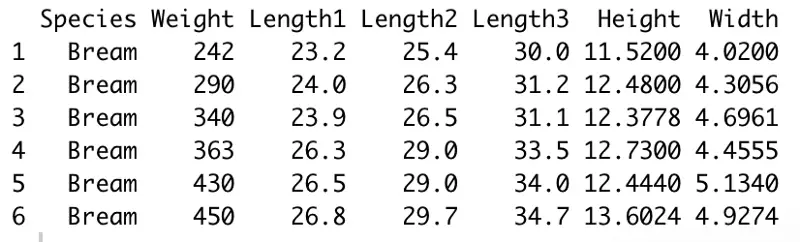
Image by author
Awesome! Everything seems to be working fine, so we can proceed with the basic exploratory data analysis.
Basic EDA
The first thing we need to do is to check for missing values. The process is as simple as with Python:
any(is.na(df))
Executing this code yields a big uppercase FALSE in the console, indicating there are no missing values — so we can proceed with the analysis.
Let’s make a couple of visualization next. Nothing complex, just to get the feel of how our dataset behaves. The first visualization is a scatter plot of fish weight vs height, colored by the fish species. This code snippet does the trick:
ggplot(data=df, aes(x=Weight, y=Height)) +
geom_point(aes(color=Species, size=10, alpha=0.7))
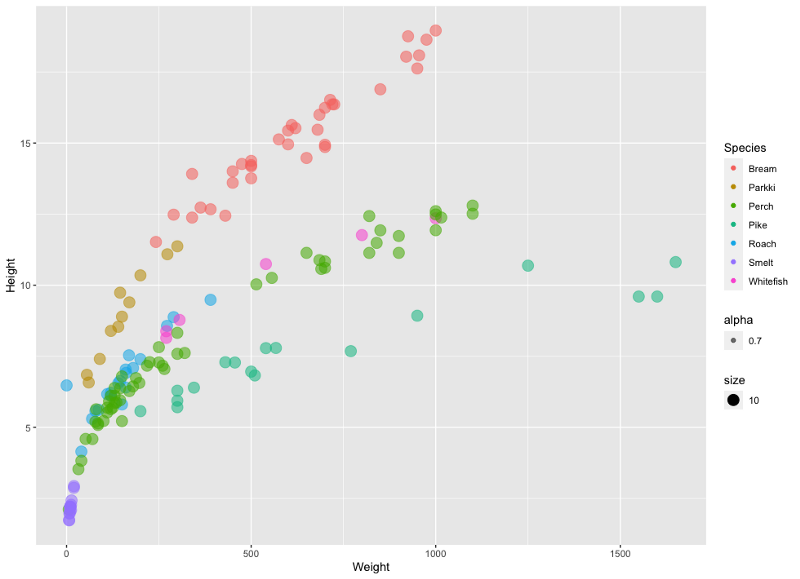
Image by author
Just from the color, we can see the fish species are nicely separated (in most cases). Next, let’s check for the correlation between the attributes. Here, the corrgram library comes in handy. Here’s the code:
corrgram(df, lower.panel=panel.shade, upper.panel=panel.cor)
And here’s the plot:
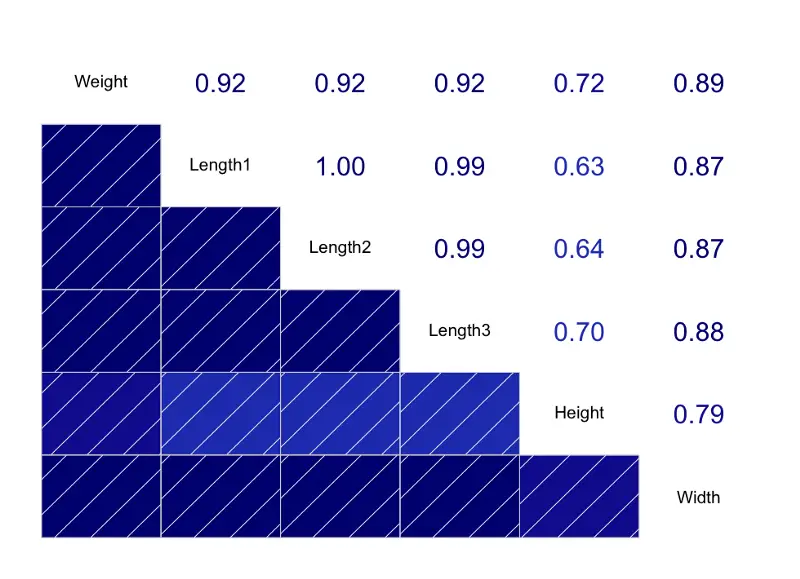
Image by author
The correlations are insanely high between almost all attributes. But we won’t do anything about it, since the aim of this article is to discuss linear regression and not the exploratory data analysis.
Keep in mind — this will most likely result in model overfitting, but more on that later.
That concludes the EDA part, and we can continue with the actual modeling.
Model training and evaluation
This will be the longest section thus far, so get yourself a cup of coffee. We’ll start with the train/test split. We want to split our dataset into two parts, one (bigger) on which the model is trained, and the other (smaller) that is used for model evaluation.
Before we do anything, let’s set a random seed. Train/test split is a random process, and seed ensures the randomization works the same on yours and my computer:
set.seed(42)
Great! Let’s perform the split now. 70% of the data is used for training, and the remaining 30% is used for testing. Here’s the code:
sampleSplit <- sample.split(Y=df$Weight, SplitRatio=0.7)
trainSet <- subset(x=df, sampleSplit==TRUE)
testSet <- subset(x=df, sampleSplit==FALSE)
After executing the code, you should see two additional variables created in the top right panel:
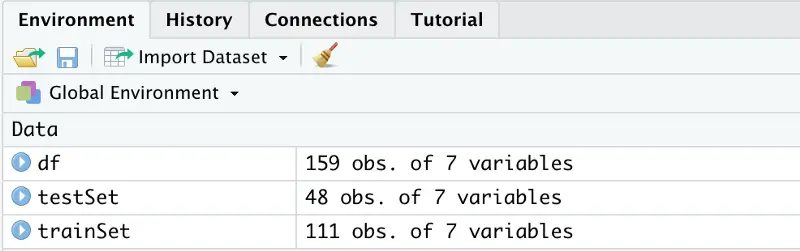
Image by author
So, we have 159 rows in total, of which 111 were allocated for model training, and the remaining 48 are used to test the model.
We can now proceed with the model training.
R uses the following syntax for linear regression models:
model <- lm(target ~ var_1 + var_2 + ... + var_n, data=train_set)
That’s okay, but imagine we had 100 predictors, then it would be a nightmare to write every single one to the equation. Instead, we can use the following syntax:
model <- lm(target ~. , data=train_set)
Keep in mind — this only works if you decide to use all predictors for model training. Accordingly, we can train the model like this:
model <- lm(formula=Weight ~ ., data=trainSet)
In a nutshell — we’re trying to predict the Weight attribute as a linear combination of every other attribute. R also handles the categorical attributes automatically. Take that, Python!
Next, we can take a look at the summary of our model:
summary(model)
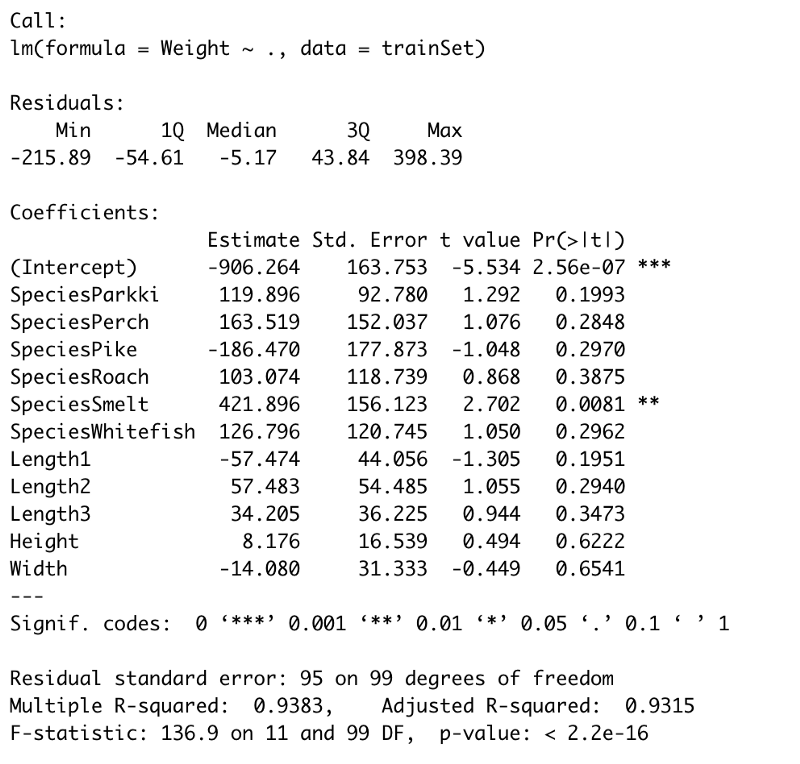
Image by author
The most interesting thing here is the P-values, displayed in the Pr(>|t|)column. Those values display the probability of a variable not being important for prediction. It’s common to use a 5% significance threshold, so if a P-value is 0.05 or below we can say that there’s a low chance it is not significant for the analysis. Sorry for the negation, that’s just how the hypotheses are formed.
Awesome! Next, we can make a residuals plot, or residuals histogram to be more precise. Here we expect to see something approximately normally distributed. Let’s see how the histogram looks like:
modelResiduals <- as.data.frame(residuals(model))
ggplot(modelResiduals, aes(residuals(model))) +
geom_histogram(fill='deepskyblue', color='black')
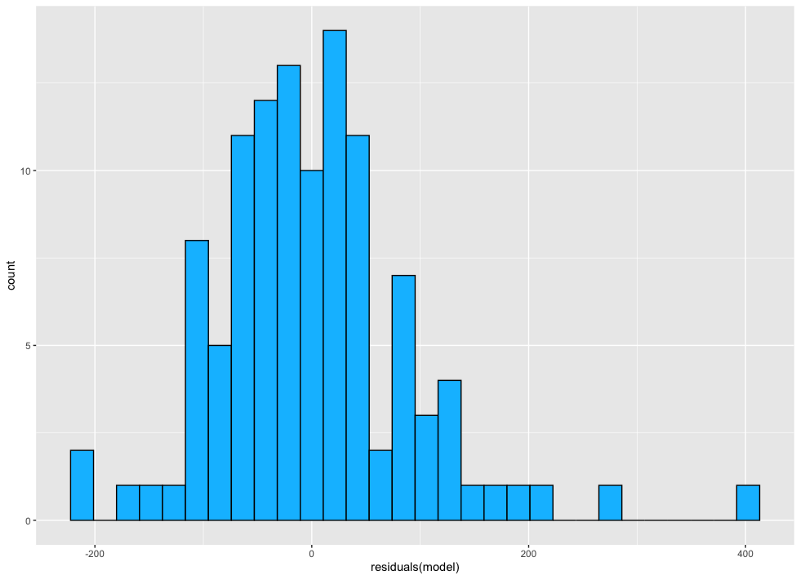
Image by author
Well, there’s a bit of skew due to the value on the far right, but after eyeballing it we can conclude that the residuals are approximately normally distributed.
And now we can finally make predictions! It’s quite easy to do so:
preds <- predict(model, testSet)
And now we can evaluate. We’ll create a dataframe of actual and predicted values, for starters:
modelEval <- cbind(testSet$Weight, preds)
colnames(modelEval) <- c('Actual', 'Predicted')
modelEval <- as.data.frame(modelEval)
Here’s how the first couple of rows look like:
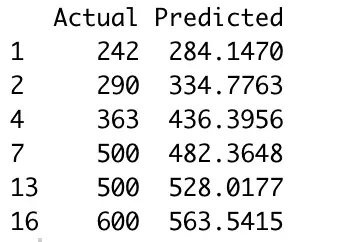
Image by author
It’s not the best model — at least not without any tuning, but we’re still getting decent results. How decent? Well, that’s what metrics like MSE and RMSE will tell us. Here are the calculations:
mse <- mean((modelEval$Actual - modelEval$Predicted)²)
rmse <- sqrt(mse)
We got the RMSE value of 95.9, and MSE is well, a square of that. This means that we’re on average wrong by 95.9 units of Weight. I’ll leave it up to you to decide how good or bad that is.
In my opinion, the model has overfitted on the training data, due to large correlation coefficients between the input variables. Also, the coefficient of determination value (R2) for the train set is insanely high (0.93+).
And that’s just enough for today. Let’s wrap things up in the next section.
Before you go
I hope this article was easy enough to follow along. We covered the simplest machine learning algorithm and touched a bit on exploratory data analysis. It’s a lot to digest for a single article, I know, but the topic isn’t that hard.
We’ll cover logistic regression next, approximately in 3–4 days, so stay tuned if that’s something you find interesting.
Thanks for reading.

